!which pip~/projects/data/Brickman/conda/envs/scvi-1.0.0/bin/pip!which pip~/projects/data/Brickman/conda/envs/scvi-1.0.0/bin/pipimport scvi
import pandas as pd
import scanpy as sc
import numpy as np
import matplotlib.pyplot as plt
from rich import print
from scib_metrics.benchmark import Benchmarker
from scvi.model.utils import mde
import warnings
from lightning_fabric.plugins.environments.slurm import PossibleUserWarning
warnings.simplefilter(action='ignore', category=PossibleUserWarning)
warnings.simplefilter(action='ignore', category=UserWarning)
warnings.simplefilter(action='ignore', category=FutureWarning)
scvi.settings.seed = 42/home/fdb589/projects/data/Brickman/conda/envs/scvi-1.0.0/lib/python3.10/site-packages/scvi/_settings.py:63: UserWarning: Since v1.0.0, scvi-tools no longer uses a random seed by default. Run `scvi.settings.seed = 0` to reproduce results from previous versions.
self.seed = seed
/home/fdb589/projects/data/Brickman/conda/envs/scvi-1.0.0/lib/python3.10/site-packages/scvi/_settings.py:70: UserWarning: Setting `dl_pin_memory_gpu_training` is deprecated in v1.0 and will be removed in v1.1. Please pass in `pin_memory` to the data loaders instead.
self.dl_pin_memory_gpu_training = (
/home/fdb589/projects/data/Brickman/conda/envs/scvi-1.0.0/lib/python3.10/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
[rank: 0] Global seed set to 42sc.set_figure_params(figsize=(10, 6))
%config InlineBackend.print_figure_kwargs={'facecolor' : "w"}
%config InlineBackend.figure_format='retina'plt.rcParams['svg.fonttype'] = 'none'adata = sc.read("../data/processed/32_human_adata.h5ad")
adataAnnData object with n_obs × n_vars = 2323 × 62754
obs: 'day', 'ct', 'experiment', 'technology', 'n_counts', 'n_genes', 'ct_fine'
layers: 'counts'adata.obs.experiment = adata.obs.experiment.str.replace('_', ' et al., ').astype('category')
adata.obs['batch'] = adata.obs.experiment
adata.obs['stage'] = adata.obs.ct.cat.rename_categories({
'Epiblast': 'EPI',
'Inner Cell Mass': 'ICM',
'Primitive Endoderm': 'PrE',
'Trophectoderm': 'TE'
}).cat.reorder_categories(['Oocyte', 'Pronucleus', 'Zygote', '2C', '4C', '8C', 'Morula', 'TE', 'ICM', 'EPI', 'PrE', 'Unknown'])
adata.obs['timepoint'] = adata.obs.ct.astype(str)
timepoint_mask = adata.obs.ct.isin(['Unknown', 'Trophectoderm', 'Inner Cell Mass', 'Primitive Endoderm', 'Epiblast'])
adata.obs.loc[timepoint_mask, 'timepoint'] = 'E' + adata.obs.loc[timepoint_mask, 'day'].astype(str)
adata.obs.ct = adata.obs.ct.astype('category')adata.obs['ct_orig'] = adata.obs.ct
adata.obs.ct = adata.obs.ct_fineENSG_to_SYMBOL = pd.read_csv('../data/external/human/Homo_sapiens.GRCh38.110.ENSG_to_SYMBOL.tsv', delimiter=" ", header=None)
ENSG_to_SYMBOL.columns = ['ensembl','symbol']
ENSG_to_SYMBOL_noName = pd.read_csv('../data/external/human/Homo_sapiens.GRCh38.110.ENSG_to_SYMBOL_noName.tsv', delimiter=" ", header=None)
nameless_df = pd.DataFrame(
data = {
'ensembl' : list(set(ENSG_to_SYMBOL_noName[0].tolist()) - set(ENSG_to_SYMBOL.ensembl.tolist())),
'symbol' : list(set(ENSG_to_SYMBOL_noName[0].tolist()) - set(ENSG_to_SYMBOL.ensembl.tolist())),
})
ENSG_to_SYMBOL = pd.concat([ENSG_to_SYMBOL, nameless_df])
ENSG_to_SYMBOL.set_index('ensembl', inplace=True)adata.var['symbol'] = ENSG_to_SYMBOL.loc[adata.var_names, 'symbol']# remove mitochondrial genes
adata = adata[:, adata.var[~adata.var.symbol.str.startswith('MT-')].index].copy()
# remove ribosomal genes
adata = adata[:, adata.var[~adata.var.symbol.str.startswith(('RPS', 'RPL'))].index].copy()# sc.pl.highest_expr_genes(adata, n_top=20)# adata.uns['log1p']["base"] = None
sc.pp.highly_variable_genes(
adata,
flavor="cell_ranger",
n_top_genes=3_000,
batch_key="batch",
subset=True,
)
adata.shape(2323, 3000)# sc.pp.highly_variable_genes(
# adata,
# flavor="seurat_v3",
# n_top_genes=3_000,
# layer="counts",
# batch_key="batch",
# subset=True,
# )
# adata.shapeimport jax
jax.devices()[gpu(id=0), gpu(id=1), gpu(id=2), gpu(id=3)]scvi.model.SCVI.setup_anndata(
adata,
layer="counts",
batch_key="batch",
)
vae = scvi.model.SCVI(adata, n_layers=2, gene_likelihood='nb')
vaeSCVI Model with the following params: n_hidden: 128, n_latent: 10, n_layers: 2, dropout_rate: 0.1, dispersion: gene, gene_likelihood: nb, latent_distribution: normal Training status: Not Trained Model's adata is minified?: False
vae.train(use_gpu=1, max_epochs=400, early_stopping=True)GPU available: True (cuda), used: True
TPU available: False, using: 0 TPU cores
IPU available: False, using: 0 IPUs
HPU available: False, using: 0 HPUs
LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0,1,2,3]
`Trainer.fit` stopped: `max_epochs=400` reached.Epoch 400/400: 100%|██████████████| 400/400 [00:58<00:00, 6.87it/s, v_num=1, train_loss_step=6.95e+3, train_loss_epoch=6.45e+3]Epoch 400/400: 100%|██████████████| 400/400 [00:58<00:00, 6.83it/s, v_num=1, train_loss_step=6.95e+3, train_loss_epoch=6.45e+3]pd.concat([vae.history['elbo_train'], vae.history['elbo_validation']], axis=1).plot.line(marker='o')<Axes: xlabel='epoch'>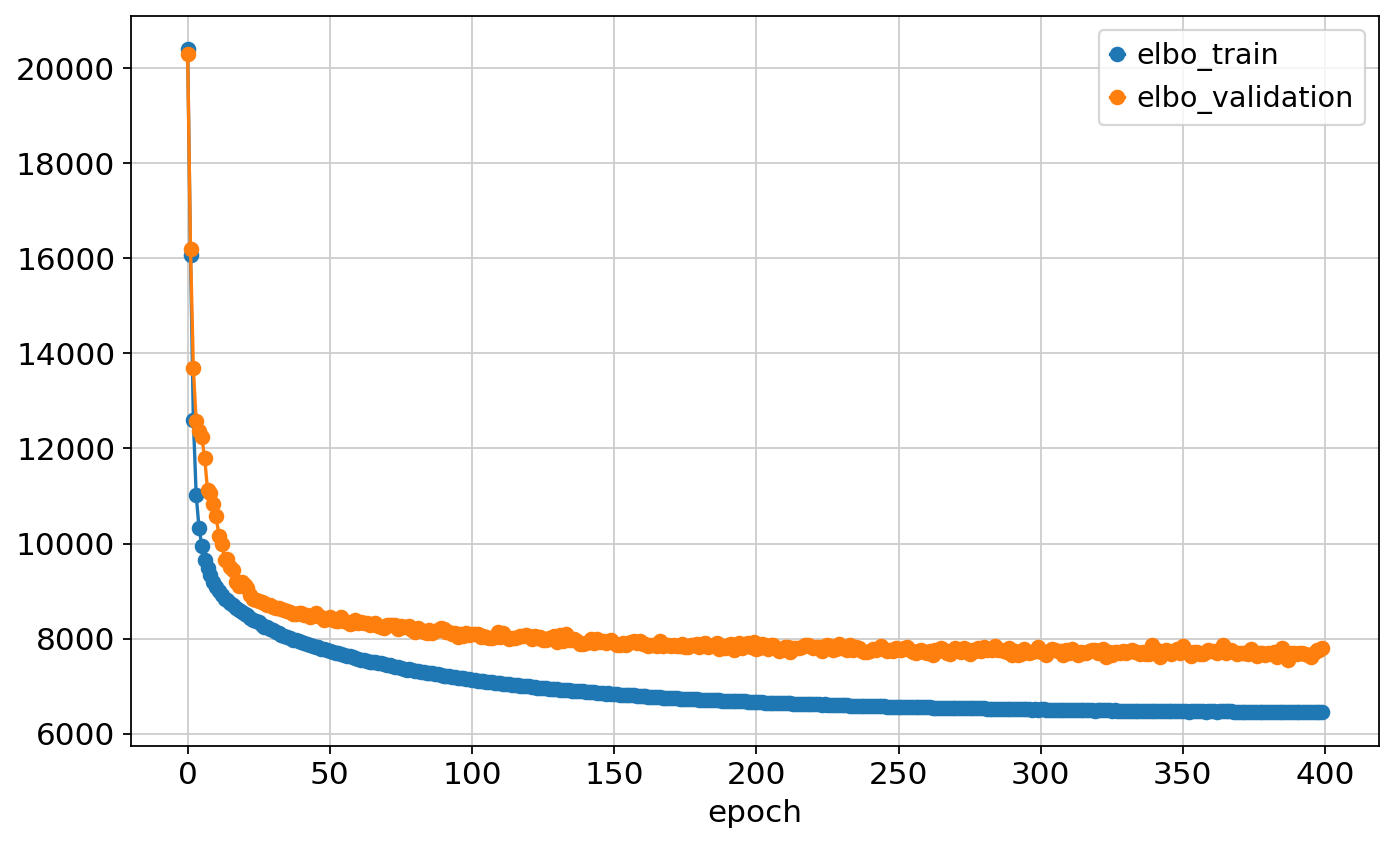
# fig, ax = plt.subplots(1, 12, figsize=[25, 4])
# for idx, key in enumerate(vae.history.keys()):
# vae.history[key].plot(title=key, ax=ax[idx])adata.obsm["X_scVI"] = vae.get_latent_representation(adata)
adata.obsm["X_mde_scVI"] = mde(adata.obsm["X_scVI"])
adata.layers['scVI_normalized'] = vae.get_normalized_expression(return_numpy=True)vae.save("../results/02_human_integration/scvi", overwrite=True, save_anndata=True)lvae = scvi.model.SCANVI.from_scvi_model(
vae,
adata=adata,
labels_key="ct",
unlabeled_category="Unknown",
)
lvaeScanVI Model with the following params: unlabeled_category: Unknown, n_hidden: 128, n_latent: 10, n_layers: 2, dropout_rate: 0.1, dispersion: gene, gene_likelihood: nb Training status: Not Trained Model's adata is minified?: False
max_epochs_scanvi = int(np.min([10, np.max([2, round(200 / 3.0)])]))
print(max_epochs_scanvi)
lvae.train(max_epochs=15)10
INFO Training for 15 epochs.
Epoch 15/15: 100%|███████████████████| 15/15 [00:05<00:00, 2.91it/s, v_num=1, train_loss_step=6.7e+3, train_loss_epoch=6.53e+3]Epoch 15/15: 100%|███████████████████| 15/15 [00:05<00:00, 2.74it/s, v_num=1, train_loss_step=6.7e+3, train_loss_epoch=6.53e+3]GPU available: True (cuda), used: True
TPU available: False, using: 0 TPU cores
IPU available: False, using: 0 IPUs
HPU available: False, using: 0 HPUs
LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0,1,2,3]
`Trainer.fit` stopped: `max_epochs=15` reached.# fig, ax = plt.subplots(3, 3, figsize=[20, 14])
# for idx, key in enumerate(lvae.history.keys()):
# lvae.history[key].plot(title=key, ax=ax[idx // 3 , idx % 3])adata.obsm["X_scANVI"] = lvae.get_latent_representation(adata)
adata.obsm["X_mde_scANVI"] = mde(adata.obsm["X_scANVI"])
adata.layers['scANVI_normalized'] = lvae.get_normalized_expression(return_numpy=True)lvae.save("../results/02_human_integration/scanvi", overwrite=True, save_anndata=True)import scgenscgen.SCGEN.setup_anndata(adata, batch_key="batch", labels_key="ct")mscgen = scgen.SCGEN(adata)mscgen.train(
max_epochs=100,
batch_size=32,
early_stopping=True,
early_stopping_patience=20,
)GPU available: True (cuda), used: True
TPU available: False, using: 0 TPU cores
IPU available: False, using: 0 IPUs
HPU available: False, using: 0 HPUs
LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0,1,2,3]Epoch 29/100: 29%|██████▉ | 29/100 [00:13<00:33, 2.10it/s, v_num=1, train_loss_step=50.1, train_loss_epoch=45]
Monitored metric elbo_validation did not improve in the last 20 records. Best score: 698.829. Signaling Trainer to stop.pd.concat([mscgen.history['elbo_train'], mscgen.history['elbo_validation']], axis=1).plot.line(marker='o')<Axes: xlabel='epoch'>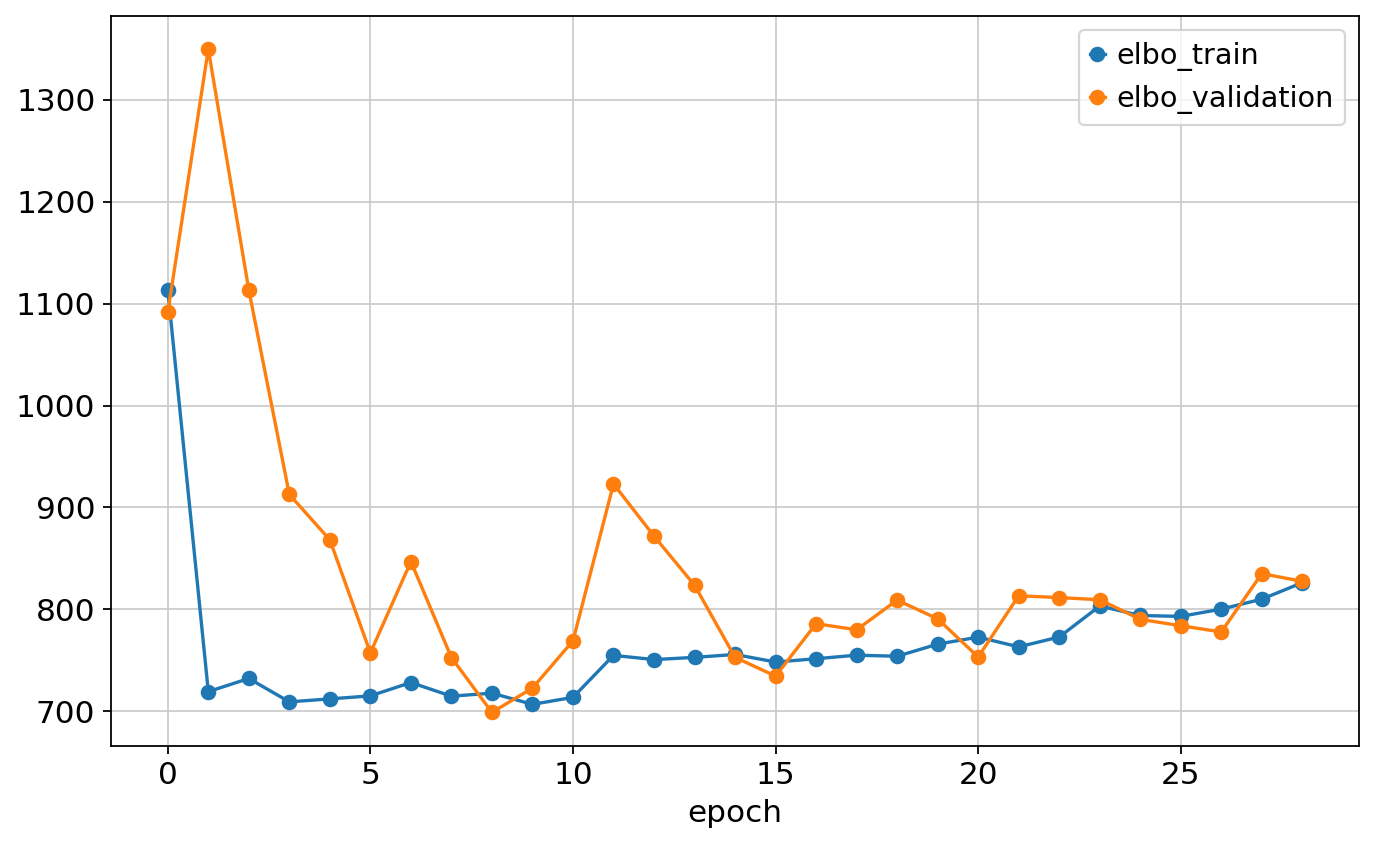
adata.obsm["X_scgen"] = mscgen.batch_removal().obsm['corrected_latent']
adata.obsm["X_mde_scgen"] = mde(adata.obsm["X_scgen"])
adata.layers['scgen_decoded_expr'] = mscgen.get_decoded_expression()INFO Input AnnData not setup with scvi-tools. attempting to transfer AnnData setup mscgen.save("../results/02_human_integration/scgen", overwrite=True, save_anndata=True)bm = Benchmarker(
adata,
batch_key="batch",
label_key="ct",
embedding_obsm_keys=["X_pca", "X_scVI", "X_scANVI", "X_scgen"],
n_jobs=-1,
)
bm.benchmark()
bm.plot_results_table(min_max_scale=False, save_dir='../results/02_human_integration/')Computing neighbors: 0%| | 0/4 [00:00<?, ?it/s]/home/fdb589/projects/data/Brickman/conda/envs/scvi-1.0.0/lib/python3.10/site-packages/umap/distances.py:1063: NumbaDeprecationWarning: The 'nopython' keyword argument was not supplied to the 'numba.jit' decorator. The implicit default value for this argument is currently False, but it will be changed to True in Numba 0.59.0. See https://numba.readthedocs.io/en/stable/reference/deprecation.html#deprecation-of-object-mode-fall-back-behaviour-when-using-jit for details.
@numba.jit()
/home/fdb589/projects/data/Brickman/conda/envs/scvi-1.0.0/lib/python3.10/site-packages/umap/distances.py:1071: NumbaDeprecationWarning: The 'nopython' keyword argument was not supplied to the 'numba.jit' decorator. The implicit default value for this argument is currently False, but it will be changed to True in Numba 0.59.0. See https://numba.readthedocs.io/en/stable/reference/deprecation.html#deprecation-of-object-mode-fall-back-behaviour-when-using-jit for details.
@numba.jit()
/home/fdb589/projects/data/Brickman/conda/envs/scvi-1.0.0/lib/python3.10/site-packages/umap/distances.py:1086: NumbaDeprecationWarning: The 'nopython' keyword argument was not supplied to the 'numba.jit' decorator. The implicit default value for this argument is currently False, but it will be changed to True in Numba 0.59.0. See https://numba.readthedocs.io/en/stable/reference/deprecation.html#deprecation-of-object-mode-fall-back-behaviour-when-using-jit for details.
@numba.jit()
/home/fdb589/projects/data/Brickman/conda/envs/scvi-1.0.0/lib/python3.10/site-packages/umap/umap_.py:660: NumbaDeprecationWarning: The 'nopython' keyword argument was not supplied to the 'numba.jit' decorator. The implicit default value for this argument is currently False, but it will be changed to True in Numba 0.59.0. See https://numba.readthedocs.io/en/stable/reference/deprecation.html#deprecation-of-object-mode-fall-back-behaviour-when-using-jit for details.
@numba.jit()
2023-12-05 19:57:29.572687: W tensorflow/compiler/tf2tensorrt/utils/py_utils.cc:38] TF-TRT Warning: Could not find TensorRT
Computing neighbors: 100%|████████████████████████████████████████████████████████████████████████| 4/4 [00:49<00:00, 12.37s/it]
Embeddings: 0%| | 0/4 [00:00<?, ?it/s]
Metrics: 0%| | 0/10 [00:00<?, ?it/s]
Metrics: 0%| | 0/10 [00:00<?, ?it/s, Bio conservation: isolated_labels]
Metrics: 10%|████▊ | 1/10 [00:02<00:20, 2.30s/it, Bio conservation: isolated_labels]
Metrics: 10%|███▍ | 1/10 [00:02<00:20, 2.30s/it, Bio conservation: nmi_ari_cluster_labels_kmeans]
Metrics: 20%|██████▊ | 2/10 [00:05<00:21, 2.64s/it, Bio conservation: nmi_ari_cluster_labels_kmeans]
Metrics: 20%|█████████▍ | 2/10 [00:05<00:21, 2.64s/it, Bio conservation: silhouette_label]
Metrics: 30%|██████████████ | 3/10 [00:05<00:12, 1.76s/it, Bio conservation: silhouette_label]
Metrics: 30%|████████████████▏ | 3/10 [00:05<00:12, 1.76s/it, Bio conservation: clisi_knn]
Metrics: 40%|█████████████████████▌ | 4/10 [00:07<00:10, 1.67s/it, Bio conservation: clisi_knn]
Metrics: 40%|██████████████████▊ | 4/10 [00:07<00:10, 1.67s/it, Batch correction: silhouette_batch]
Metrics: 50%|███████████████████████▌ | 5/10 [00:20<00:28, 5.77s/it, Batch correction: silhouette_batch]
Metrics: 50%|███████████████████████████ | 5/10 [00:20<00:28, 5.77s/it, Batch correction: ilisi_knn]
Metrics: 60%|████████████████████████████████▍ | 6/10 [00:20<00:15, 3.91s/it, Batch correction: ilisi_knn]
Metrics: 60%|█████████████████████████████▍ | 6/10 [00:20<00:15, 3.91s/it, Batch correction: kbet_per_label]
Metrics: 70%|██████████████████████████████████▎ | 7/10 [00:29<00:16, 5.43s/it, Batch correction: kbet_per_label]
Metrics: 70%|███████████████████████████████▍ | 7/10 [00:29<00:16, 5.43s/it, Batch correction: graph_connectivity]
Metrics: 80%|███████████████████████████████████████▏ | 8/10 [00:29<00:10, 5.43s/it, Batch correction: pcr_comparison]
Embeddings: 25%|████████████████████▎ | 1/4 [00:30<01:31, 30.54s/it]
Metrics: 0%| | 0/10 [00:00<?, ?it/s]
Metrics: 0%| | 0/10 [00:00<?, ?it/s, Bio conservation: isolated_labels]
Metrics: 10%|████▊ | 1/10 [00:00<00:01, 4.56it/s, Bio conservation: isolated_labels]
Metrics: 10%|███▍ | 1/10 [00:00<00:01, 4.56it/s, Bio conservation: nmi_ari_cluster_labels_kmeans]
Metrics: 20%|██████▊ | 2/10 [00:02<00:11, 1.46s/it, Bio conservation: nmi_ari_cluster_labels_kmeans]
Metrics: 20%|█████████▍ | 2/10 [00:02<00:11, 1.46s/it, Bio conservation: silhouette_label]
Metrics: 30%|████████████████▏ | 3/10 [00:02<00:10, 1.46s/it, Bio conservation: clisi_knn]
Metrics: 40%|██████████████████▊ | 4/10 [00:02<00:08, 1.46s/it, Batch correction: silhouette_batch]
Metrics: 50%|███████████████████████▌ | 5/10 [00:03<00:03, 1.45it/s, Batch correction: silhouette_batch]
Metrics: 50%|███████████████████████████ | 5/10 [00:03<00:03, 1.45it/s, Batch correction: ilisi_knn]
Metrics: 60%|█████████████████████████████▍ | 6/10 [00:03<00:02, 1.45it/s, Batch correction: kbet_per_label]
Metrics: 70%|██████████████████████████████████▎ | 7/10 [00:05<00:02, 1.25it/s, Batch correction: kbet_per_label]
Metrics: 70%|███████████████████████████████▍ | 7/10 [00:05<00:02, 1.25it/s, Batch correction: graph_connectivity]
Metrics: 80%|███████████████████████████████████████▏ | 8/10 [00:05<00:01, 1.25it/s, Batch correction: pcr_comparison]
Embeddings: 50%|████████████████████████████████████████▌ | 2/4 [00:36<00:32, 16.34s/it]
Metrics: 0%| | 0/10 [00:00<?, ?it/s]
Metrics: 0%| | 0/10 [00:00<?, ?it/s, Bio conservation: isolated_labels]
Metrics: 10%|███▍ | 1/10 [00:00<00:00, 17.03it/s, Bio conservation: nmi_ari_cluster_labels_kmeans]
Metrics: 20%|██████▊ | 2/10 [00:01<00:05, 1.38it/s, Bio conservation: nmi_ari_cluster_labels_kmeans]
Metrics: 20%|█████████▍ | 2/10 [00:01<00:05, 1.38it/s, Bio conservation: silhouette_label]
Metrics: 30%|████████████████▏ | 3/10 [00:01<00:05, 1.38it/s, Bio conservation: clisi_knn]
Metrics: 40%|██████████████████▊ | 4/10 [00:01<00:04, 1.38it/s, Batch correction: silhouette_batch]
Metrics: 50%|███████████████████████▌ | 5/10 [00:01<00:01, 3.86it/s, Batch correction: silhouette_batch]
Metrics: 50%|███████████████████████████ | 5/10 [00:01<00:01, 3.86it/s, Batch correction: ilisi_knn]
Metrics: 60%|█████████████████████████████▍ | 6/10 [00:01<00:01, 3.86it/s, Batch correction: kbet_per_label]
Metrics: 70%|██████████████████████████████████▎ | 7/10 [00:03<00:01, 1.87it/s, Batch correction: kbet_per_label]
Metrics: 70%|███████████████████████████████▍ | 7/10 [00:03<00:01, 1.87it/s, Batch correction: graph_connectivity]
Embeddings: 75%|████████████████████████████████████████████████████████████▊ | 3/4 [00:40<00:10, 10.49s/it]
Metrics: 0%| | 0/10 [00:00<?, ?it/s]
Metrics: 0%| | 0/10 [00:00<?, ?it/s, Bio conservation: isolated_labels]
Metrics: 10%|████▊ | 1/10 [00:00<00:02, 4.01it/s, Bio conservation: isolated_labels]
Metrics: 10%|███▍ | 1/10 [00:00<00:02, 4.01it/s, Bio conservation: nmi_ari_cluster_labels_kmeans]
Metrics: 20%|██████▊ | 2/10 [00:02<00:11, 1.45s/it, Bio conservation: nmi_ari_cluster_labels_kmeans]
Metrics: 20%|█████████▍ | 2/10 [00:02<00:11, 1.45s/it, Bio conservation: silhouette_label]
Metrics: 30%|████████████████▏ | 3/10 [00:02<00:10, 1.45s/it, Bio conservation: clisi_knn]
Metrics: 40%|██████████████████▊ | 4/10 [00:02<00:08, 1.45s/it, Batch correction: silhouette_batch]
Metrics: 50%|███████████████████████▌ | 5/10 [00:03<00:03, 1.40it/s, Batch correction: silhouette_batch]
Metrics: 50%|███████████████████████████ | 5/10 [00:03<00:03, 1.40it/s, Batch correction: ilisi_knn]
Metrics: 60%|█████████████████████████████▍ | 6/10 [00:03<00:02, 1.40it/s, Batch correction: kbet_per_label]
Metrics: 70%|██████████████████████████████████▎ | 7/10 [00:04<00:01, 1.54it/s, Batch correction: kbet_per_label]
Metrics: 70%|███████████████████████████████▍ | 7/10 [00:04<00:01, 1.54it/s, Batch correction: graph_connectivity]
Metrics: 80%|███████████████████████████████████████▏ | 8/10 [00:04<00:01, 1.54it/s, Batch correction: pcr_comparison]
Embeddings: 100%|█████████████████████████████████████████████████████████████████████████████████| 4/4 [00:46<00:00, 11.55s/it]
INFO Late epiblast consists of a single batch or is too small. Skip.
INFO Trophectoderm_10.0 consists of a single batch or is too small. Skip.
INFO Trophectoderm_8.0 consists of a single batch or is too small. Skip.
INFO Trophectoderm_9.0 consists of a single batch or is too small. Skip.
INFO Late epiblast consists of a single batch or is too small. Skip.
INFO Trophectoderm_10.0 consists of a single batch or is too small. Skip.
INFO Trophectoderm_8.0 consists of a single batch or is too small. Skip.
INFO Trophectoderm_9.0 consists of a single batch or is too small. Skip.
INFO Late epiblast consists of a single batch or is too small. Skip.
INFO Trophectoderm_10.0 consists of a single batch or is too small. Skip.
INFO Trophectoderm_8.0 consists of a single batch or is too small. Skip.
INFO Trophectoderm_9.0 consists of a single batch or is too small. Skip.
INFO Late epiblast consists of a single batch or is too small. Skip.
INFO Trophectoderm_10.0 consists of a single batch or is too small. Skip.
INFO Trophectoderm_8.0 consists of a single batch or is too small. Skip.
INFO Trophectoderm_9.0 consists of a single batch or is too small. Skip. 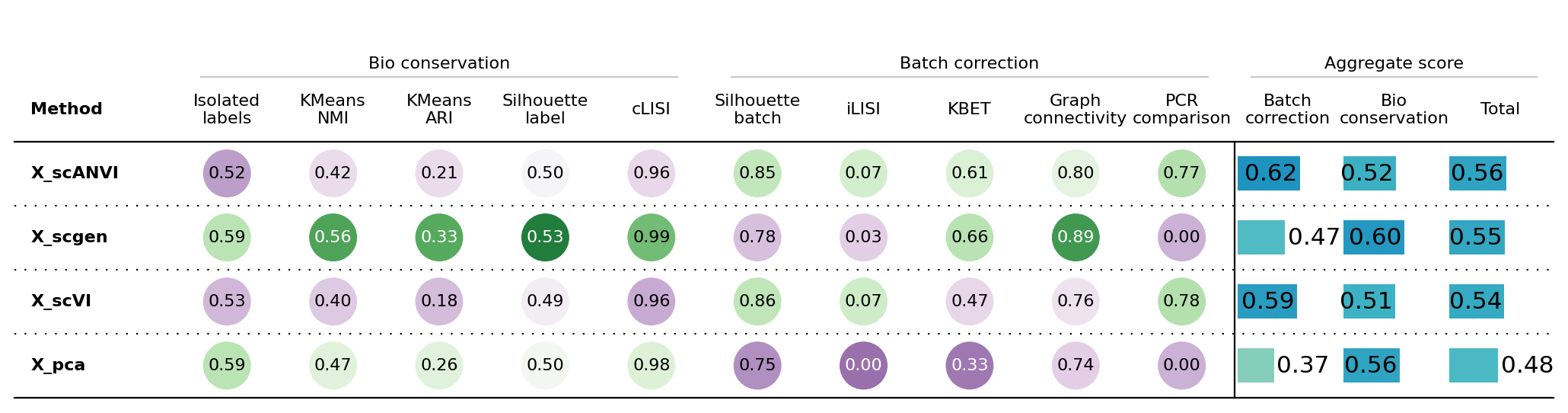
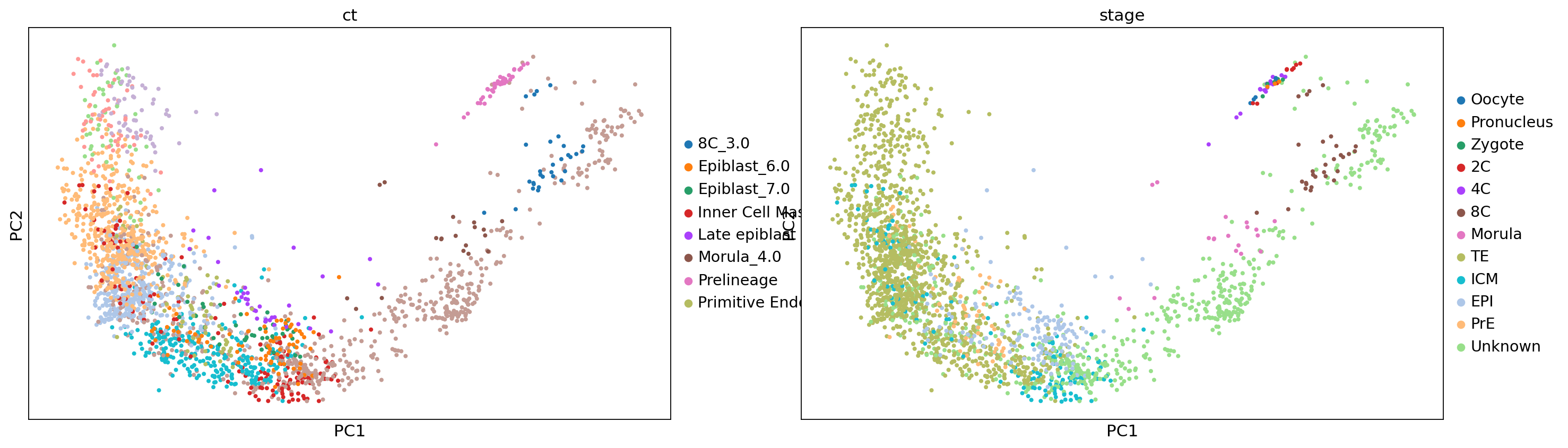
<plottable.table.Table at 0x7f5f707142e0>sc.pl.pca(adata, color=['ct', 'stage'])
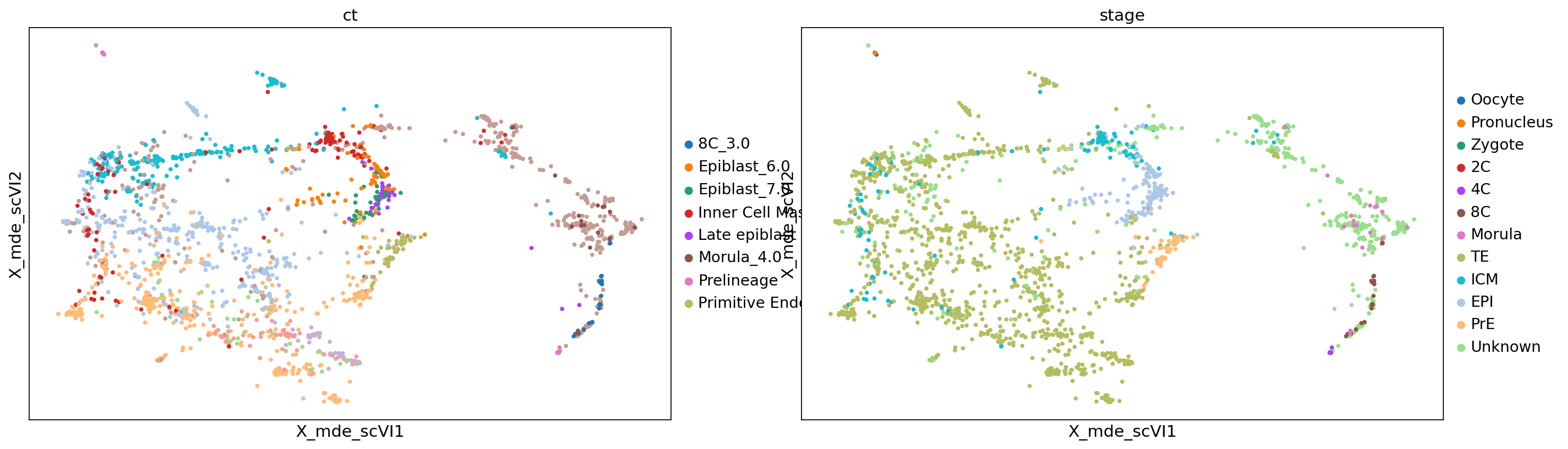
sc.pl.embedding(adata, color=['ct', 'stage'], basis='X_mde_scVI')
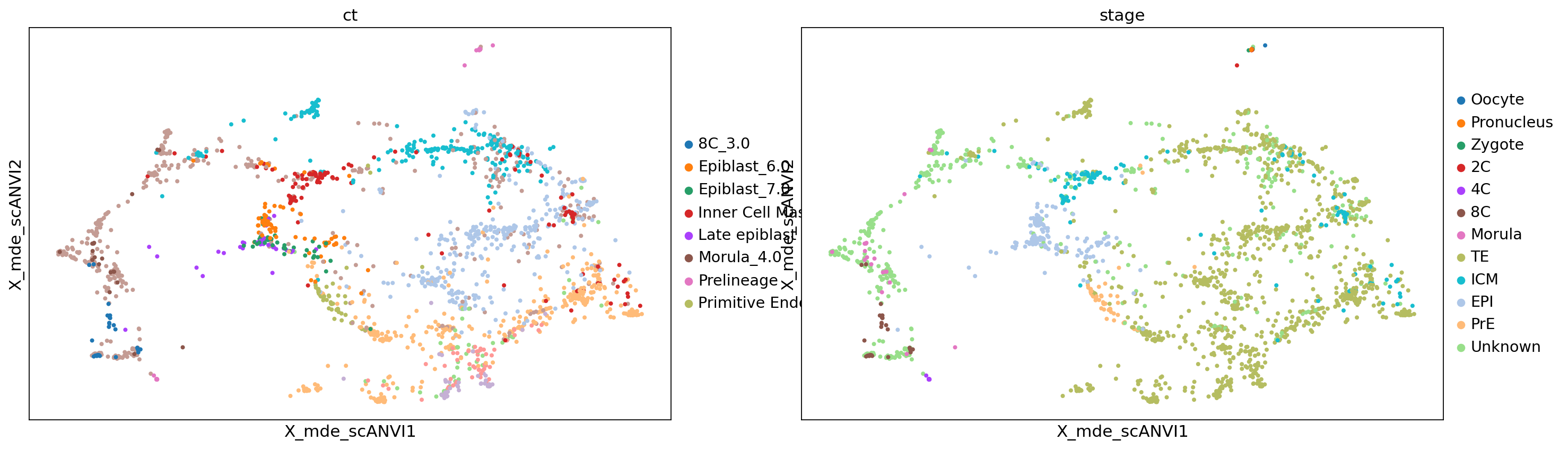
sc.pl.embedding(adata, color=['ct', 'stage'], basis='X_mde_scANVI')
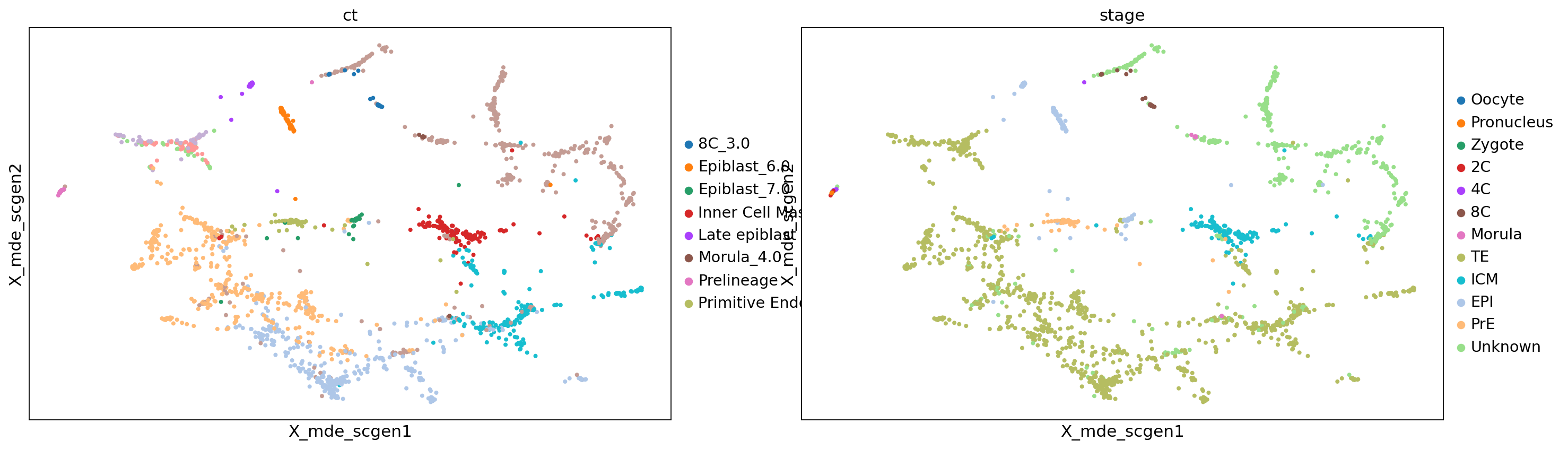
sc.pl.embedding(adata, color=['ct', 'stage'], basis='X_mde_scgen')